VERVENet capabilities to go viral
My Bioinformatics Cookbook
OpenCloud for open science
Groups in protocols.io
Now that we have ironed out most of the bugs through the VERVE Net, we are opening up the group functionality in protocols.io. This means that you can create your own group (for a lab or class) and enter protocols associated with that group. You can reach groups through the regular protocols.io site here. You will then click on the ‘explore’ button and go into ‘browse groups.’ You can create a group by clicking the “+ groups” sign next to your picture in the upper left hand corner of protocols.io. *Be sure to create a login first.
You can also join the viral ecology group: VERVE Net. Click on the ‘join group’ button on the VERVE Net group page.
UA News: Go Viral
 Check out the recent UA news article on our VERVE Net grant. Also, our first implementation of VERVE Net is up and running check it out.
Check out the recent UA news article on our VERVE Net grant. Also, our first implementation of VERVE Net is up and running check it out.
The quest for wound diagnostics

Announcing VERVE Net
VERVE Net: Viral Ecology Research and Virtual Exchange Network
Bonnie Hurwitz, PhD, PI, University of Arizona; Lenny Teytelman, PhD, Scientific Lead, ZappyLab
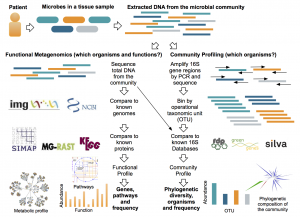
The advent of metagenomic methods to sequence DNA directly from an environment has revolutionized viral ecology, making it possible to “see” natural viral communities that could not be previously studied through culture. This technological leap has enabled further innovation in: extracting and sequencing limited viral DNA from communities, enriching and sequencing wild viruses through single-cell genomics, and creating new bioinformatics methods for large-scale comparative and functional metagenomics. Yet, the knowledge for specialized techniques in viral ecology remains in a subset of labs. We are developing a viral ecology community forum called VERVE Net to increase connectivity and knowledge dissemination in viral ecology research at all levels from undergraduates to accomplished viral ecologists. VERVE Net will leverage and refine existing software from ZappyLab to enhance a researcher’s ability to: discuss and share protocols (via protocols.io), connect with fellow community members (VERVE Net community forum), and learn about new and innovative research in the field (via PubChase). In delivering these valuable tools, VERVE Net will become a central resource to connect, collaborate, share and innovate for the viral ecology community.
The latest on iMicrobe